Like finding a needle in a haystack, Liviu Movileanu can find a single molecule in blood. The new technology, developed by Movileanu and Avinash Thakur, has wide‐ranging applications from diagnostic tests to drug discovery. Liviu Movileanu, physics professor in the College of Arts and Sciences (A&S) and Avinash Thakur, a recent doctoral graduate, presented their research at the Annual Meeting of the Biophysical Society on February 17 in San Diego, California.

Movileanu began “chasing protein‐protein interactions under disease‐like conditions” after his postdoctoral studies. As a postdoc, he began using nanopores, tiny holes in cell membranes through which scientists shoot electrical currents. When an individual molecule, like a protein, enters the pore the electrical current changes in a way that allows scientists to identify the molecule’s identity. But in order to understand how different proteins interacted with each other, he needed to modify the system. One of the challenges was that grouped proteins are too big to fit into the nanopore, where the measurement typically takes place. To overcome this, Thakur and Movileanu went fishing.
Thakur and Movileanu created molecular “fishing” by fusing a modified receptor that acts like a “hook and bait,” via a short flexible protein “line,” to a protein nanopore “rod and reel.” Then they added an extra little protein that acts like a fishing “bobber.” When there is nothing on the “hook” it bobs rapidly into and out of the nanopore. Yet, when something grabs hold it stops moving around, which alerts scientists that there’s something on the “hook.” This ingenious approach enabled them to use a pore that’s too small for big protein complexes to become a sensor for protein interactions.
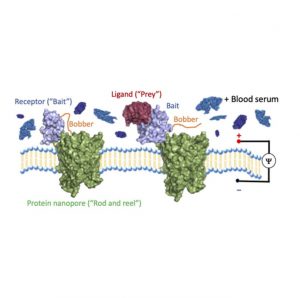
Unlike fishing bait, where a worm might catch a trout or a catfish, Thakur and Movileanu’s bait is both extremely specific and totally customizable to find any protein of interest, and it even works in complex solutions like blood or biopsy samples. This precise protein engineering has practical significance in diagnostics, and because of its specificity there are no potential false‐positive signals produced by the constituents of a complex biofluid sample. Additionally, calculations with several of these “fishing rods” can reveal the concentration of the protein of interest in the solution.
“This sensor has realistic prospects in many biomedical areas,” Movileanu says. It could be scaled up to include many proteins for diagnosing diseases, discovering disease biomarkers, or finding new drugs that disrupt or enhance protein interactions. “We demonstrated the proof of concept,” said Movileanu, and the next step is scaling it up to find the needles in a lot more haystacks.
This article was prepared by the Biophysical Society.
